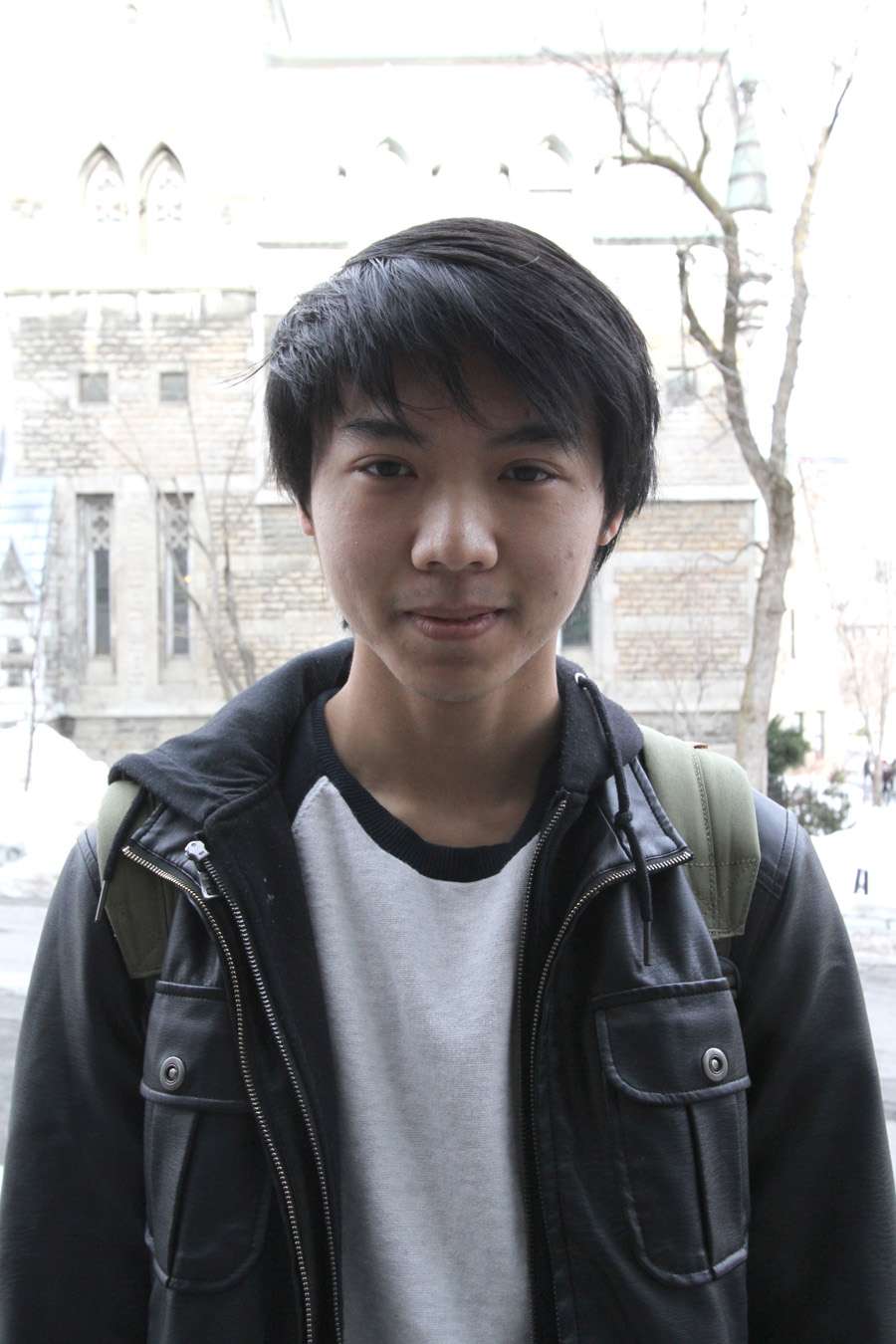
C. elegans, more formally known as Caenorhabditis elegans, is a simple, transparent roundworm often used in genetic research. After working with the organism from a neuroscience perspective under the supervision of associate professor Joseph Dent, U2 interdepartmental honours student Daegan Sit combined his experience with the worm and his interest in immunology through his project in the Ausubel Lab at the Massachusetts General Hospital.
“My project was basically about C. elegans immunology,” Sit explained. “C. elegans is not normally known [in] immunology, and that is why I wanted to work in this lab. They are one of the first groups to try using this simple organism to study immunology.”
C. elegans is such a simplistic organism that it does not possess an adaptive immune system—one of two critical components of the overall immune response—which is why many labs have not explored research in immunology with this roundworm. However, the group at the Massachusetts General Hospital found that some pathogens that infected humans also infected these worms, leading the scientists to continue research on the organism. The main pathogen that the researchers worked with was the bacteria Pseudomonas aeruginosa, a bacterium responsible for infections of burn injuries and of the outer ear, among other diseases.
“[The lab] decided to try using [C. elegans] in high throughput studies [where researchers quickly conduct thousands of tests], and that became very effective because you can grow so many [worms],” said Sit. “[The worms] basically just eat [the bacterium] E. coli; so it is relatively cheap to produce hundreds of thousands of these worms.”
Sit’s project dealt specifically with designing a method to perform the high throughput screening experiments that the lab was interested in conducting. He worked with a library of mutant Pseudomonas aeruginosa, each of which had a different gene that was missing from their DNA.
“My job was to design a way to infect all of the worms with the different units of bacteria and see which strains of bacteria did not kill the worms as well,” explained Sit. “Using the life of the worms as a readout, we could tell what genes are important in the Pseudomonas aeruginosa.”
Through this experiment, Sit was able to determine which bacteria were not as effective at killing the C. elegans when the worms were infected with the Pseudomonas aeruginosa. The lab could then look at what gene the Pseudomonas aeruginosa was missing to see which genes were important in its pathogenicity.
According to Sit, the greatest technical challenge in designing this screening method was getting the bacteria to grow at the same rate.
“Because they’re mutants […] some strains of Pseudomonas aeruginosa tend to grow at different rates,” Sit said. “Also, sometimes the bacteria would start growing from a different concentration. To solve this problem I first troubleshooted a variety of different growing and incubation techniques to try and get all strains to grow at a comparable rate. As well, I would keep track of the final density of the bacteria right before they were used for infection. When we would find out whether or not the bacteria killed the C. elegans, we would look back to see whether the density of bacteria was extremely low. If this was the case, it could tell us that perhaps the mutant bacteria did not kill the worms because they grew poorly and not because the mutant had a defect in its pathogenesis, which is what we were looking for.”.”

Sit developed this technique through trouble shooting, as well as by looking into the past about how other researchers had solved problems in growing bacteria in a high throughput environment.
While Sit has worked in numerous other labs, his experience at the Oswald Lab provided him with the opportunity to gain independence in his research.
“I worked under a postdoctorate researcher, Dr. Kirienko, who taught me all the relevant techniques and gave great guidance; but she really pushed me to work on my own, which was really important for my own development,” Sit said.
Sit’s efforts paid off this past November, when he was awarded a second-place prize for his project at the Undergraduate Research Conference held at McGill. Sit plans to continue his involvement in research at McGill, where he is currently working for professor Nahum Sonenberg. He hopes to make the most of the mobility afforded to undergraduate researchers by taking advantage of the opportunity to work in a variety of labs before he graduates.
What is your favourite part of working in a lab?
“I think my favourite part about biological techniques is the moment when you get the data out—the read out—because it is always exciting to see whether it worked, or it is disappointing when you see everything failed. [To see the C. elegans mortality rate], I dyed the organism with a stain known as SYTOX orange, which selectively enters dead C. elegans worms [and makes them] glow. I would put the entire plate under a microscope and take a [picture] of a florescent image and just a regular image. There was this program called Cell Profiling, and it is able to calculate the area of the C. elegans worms both in the fluorescent image and the non-fluorescent image, [giving us] a readout of the percent dead.”
Advice for other students applying to a lab?
“The trick is to apply to labs when they’re not as busy or when they’re not filled up. I remember the first time I applied for a lab during the summer I [did so] kind of late—towards January, February, and March. By that time, some laboratories are filled up. It doesn’t mean you’re not qualified, but it may mean that the [lab] does not have space. I think that’s one big thing, trying to apply early on and trying to find work that you’re really interested in.”
If you could have any superhero power, what would it be?
“I think definitely reversing time would be the most useful; [a] time turner—that would be nice.”